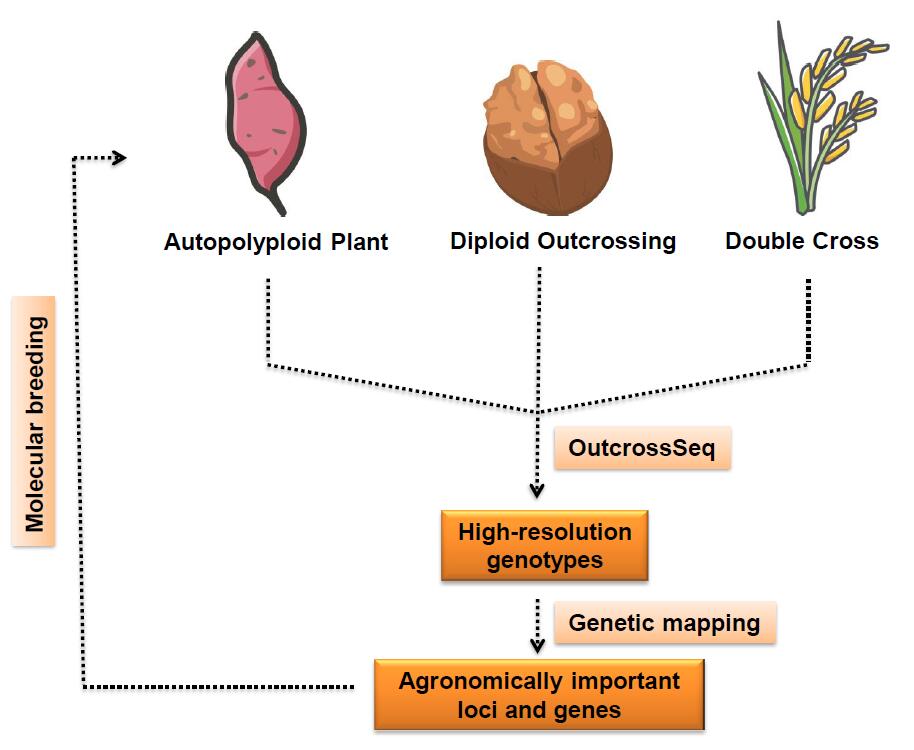
'OutcrossSeq' for identification of agronomically important genes in outcrossing crops
|
|
|
|
Any questions or suggestions, please contact
Xuehui Huang Lab xhhuang@shnu.edu.cn 100 Guilin Road, Building #14, 907 Last update: 2020/12 © 2019-2021 xhhuanglab.cn. All rights reserved. 沪ICP备19028236号. |